Crystal breakage of mechanically labile protein crystals in the mechanical solid-liquid separation
- Contact:
- Project Group:
DiSPBiotech
- Funding:
DFG
Project description
Recent progresses in the biotechnology industries have led to much higher product yields after the fermentation. In the case of proteins the purification of such high titers have become a challenging task. The common method is to use several chromatography steps to separate the protein with high purity. However, the high concentration of protein in the mother liquor requires large amounts of toxic and explosive organic solvents and large chromatography columns. Hence, another approach like selective crystallization to separate the protein from the mother liquor is becoming increasingly interesting for the pharmaceutical and biotechnology industry. Crystallized proteins not only offer advantages in the downstream processing but also have other positive properties like extended shelf life, easier product handling and in the case of therapeutic proteins different drug release properties. After the crystallization, the next step is often a solid-liquid separation. In the case of protein crystals this separation is a challenging task because in contrast to common organic or inorganic crystals, protein crystals are much more fragile to mechanical stress. Even low pressure differences may lead to crystal breakage or crystal abrasion. Since therapeutic or enzymatic proteins are often expensive products a method to analyze the mechanical stability and the influence of crystal breakage on the cake filtration with low volumes and hence low product quantities is required.

The microcomputertomography (µCT) allows to analyze filter cakes consisting of lysozyme crystals. After the sample preparation and the measurement with the Zeiss XRadia 520 Versa computer tomograph, the reconstructed data set is evaluated with the 3D image processing software Avizo. With the device, a resolution of 700nm and a voxel size of 70nm can be achieved. This however, requires an optimal suitable sample, which often is not the case for biological materials like protein crystals. The challenging part of the analysis is to reduce the data set and to separate the crystals from the background using thresholding. Afterwards, the volume and the diameter of the crystals are measured and the crystal size distribution is calculated.
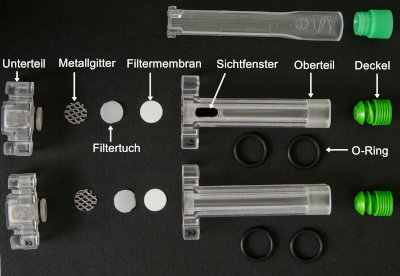 To analyze the filtration, a 3D printed filtration cell for an optical analytical laboratory centrifuge is used. This filtration device allows to measure the filter cake build up, the filtration resistance, porosity and the flux density function. The required amount of sample of about 300µl per experiment makes this setup suitable for proteins, which are still in development and hence are only available on a small scale. Material functions can be derived from the measured data, which serve as input parameters for micro and macro structure simulations. Occurring particle breakage can be quantified using optical methods before and after the filtration.
To analyze the filtration, a 3D printed filtration cell for an optical analytical laboratory centrifuge is used. This filtration device allows to measure the filter cake build up, the filtration resistance, porosity and the flux density function. The required amount of sample of about 300µl per experiment makes this setup suitable for proteins, which are still in development and hence are only available on a small scale. Material functions can be derived from the measured data, which serve as input parameters for micro and macro structure simulations. Occurring particle breakage can be quantified using optical methods before and after the filtration.
Funded by:


